Examples¶
Fitting distance metric learning algorithms¶
>>> import numpy as np
>>> from sklearn.datasets import load_iris
>>> # Loading DML Algorithm
>>> from dml import NCA
>>> # Loading dataset
>>> iris = load_iris()
>>> X = iris['data']
>>> y = iris['target']
>>> # DML construction
>>> nca = NCA()
>>> # Fitting algorithm
>>> nca.fit(X,y)
>>> # We can look at the algorithm metadata after fitting it
>>> meta = nca.metadata()
>>> meta
{'final_expectance': 0.95771240234375,
'initial_expectance': 0.8380491129557291,
'num_iters': 3}
>>> # We can see the metric the algorithm has learned.
>>> # This metric is the PSD matrix that defines how the distance is measured:
>>> # d(x,y) = (x-y).T.dot(M).dot(x-y)
>>> M = nca.metric()
>>> M
array([[ 1.19098678, 0.51293714, -2.15818151, -2.01464351],
[ 0.51293714, 1.58128238, -2.14573777, -2.10714773],
[-2.15818151, -2.14573777, 6.46881853, 5.86280474],
[-2.01464351, -2.10714773, 5.86280474, 6.83271473]])
>>> # Equivalently, we can see the learned linear map.
>>> # The distance coincides with the euclidean distance after applying the linear map.
>>> L = nca.transformer()
>>> L
array([[ 0.77961001, -0.01911998, -0.35862791, -0.23992861],
[-0.04442949, 1.00747788, -0.29936559, -0.25812144],
[-0.60744415, -0.57288453, 2.16095076, 1.35212555],
[-0.46068713, -0.48755353, 1.25732916, 2.20913531]])
>>> # Finally, we can obtain the transformed data ...
>>> Lx = nca.transform()
>>> Lx[:5,:]
array([[ 3.35902632, 2.8288461 , -1.80730485, -1.85385382],
[ 3.21266431, 2.33399305, -1.39937375, -1.51793964],
[ 3.0887811 , 2.57431109, -1.60855691, -1.64904583],
[ 2.94100652, 2.41813313, -1.05833389, -1.30275593],
[ 3.27915332, 2.93403684, -1.80384889, -1.85654046]])
>>> # ... or transform new data.
>>> X_ = np.array([[1.0,0.0,0.0,0.0],[1.0,1.0,0.0,0.0],[1.0,1.0,1.0,0.0]])
>>> Lx_ = nca.transform(X_)
>>> Lx_
array([[ 0.77961001, -0.04442949, -0.60744415, -0.46068713],
[ 0.76049003, 0.9630484 , -1.18032868, -0.94824066],
[ 0.40186212, 0.66368281, 0.98062208, 0.3090885 ]])
Similarity learning classifier extensions for Scikit-learn¶
>>> import numpy as np
>>> from sklearn.datasets import load_iris
>>> from dml import NCA, kNN, MultiDML_kNN
>>> # Loading dataset
>>> iris = load_iris()
>>> X = iris['data']
>>> y = iris['target']
>>> # Initializing transformer and predictor
>>> nca = NCA()
>>> knn = kNN(n_neighbors=7,dml_algorithm=nca)
>>> # Fitting transformer and predictor
>>> nca.fit(X,y)
>>> knn.fit(X,y)
# Now we can predict the labels for k-NN with the learned distance.
>>> knn.predict() # Also we can use predict(X_) for other datasets.
>>> # When using the training set predictions are made
>>> # leaving the sample to predict out.
array([ 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 2., 1., 2., 1., 1., 1., 1., 2.,
1., 1., 1., 1., 1., 2., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 2., 2., 2., 2.,
2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2.,
2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2.,
2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2.,
2., 2., 2., 2., 2., 2., 2.])
>>> knn.predict_proba()[-10:,:] # Again it can be used for other datasets.
array([[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0.14285714, 0.85714286],
[ 0. , 0. , 1. ],
[ 0. , 0. , 1. ],
[ 0. , 0.14285714, 0.85714286]])
>>> knn.score() # The classification score (score(X_,y_) for other datasets).
0.97333333333333338
>>> # We can also compare with the euclidean distance k-NN
>>> knn.score_orig()
0.96666666666666667
>>> # With MultiDML_kNN we can test multiple dmls. In this case, dmls are fitted automatically.
>>> lda = LDA()
>>> mknn = MultiDML_kNN(n_neighbors=7,dmls=[lda,nca])
>>> mknn.fit(X,y)
>>> # And we can predict and take scores in the same way, for every dml.
>>> # The euclidean distance will be added always in first place.
>>> mknn.score_all() # It will show [euclidean, lda, nca]
array([ 0.96666667, 0.96666667, 0.97333333])
>>> # The NCMC Classifier works like every ClassifierMixin.
>>> ncmc = NCMC_Classifier(centroids_num=2)
>>> ncmc.fit(X,y)
>>> ncmc.score(X,y)
0.95333333333333337
>>> # To learn a distance to use with NCMC Classifier, and with any other distance classifier
>>> # we can use pipelines.
>>> from sklearn.pipeline import Pipeline
>>> dml_ncmc = Pipeline([('nca',nca),('ncmc',ncmc)])
>>> dml_ncmc.fit(X,y)
>>> dml_ncmc.score(X,y)
0.97999999999999998
Plotting classifier regions induced by different distances¶
>>> import numpy as np
>>> from sklearn.datasets import load_iris
>>> from dml import NCA, LDA, NCMC_Classifier, classifier_plot, dml_plot, knn_plot,
>>> dml_multiplot, knn_pairplots
>>> # Loading dataset
>>> iris = load_iris()
>>> X = iris['data']
>>> y = iris['target']
>>> # Initializing transformers and predictors
>>> nca = NCA()
>>> lda = LDA()
>>> ncmc = NCMC_Classifier(centroids_num=2)
>>> # We can plot regions for different classifiers
>>> f1 = classifier_plot(X[:,[0,1]],y,clf=ncmc,title = "NCMC Classification",
>>> cmap="rainbow",figsize=(12,6))
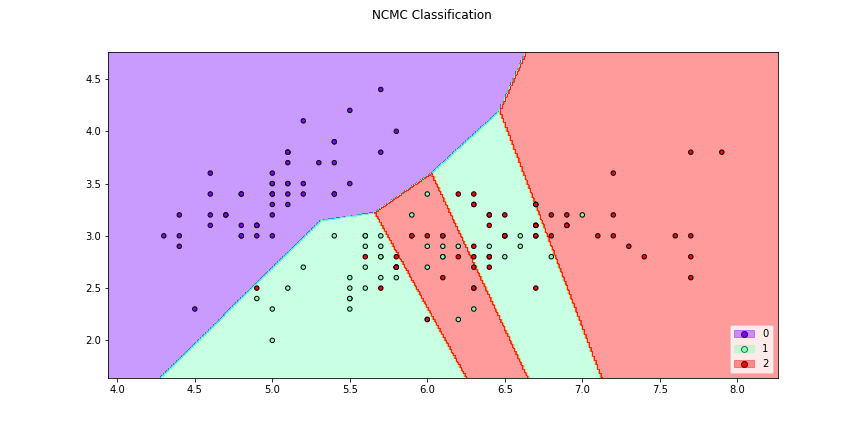
>>> f2 = knn_plot(X[:,[0,1]],y,k=3,title = "3-NN Classification", cmap="rainbow",
>>> figsize=(12,6))
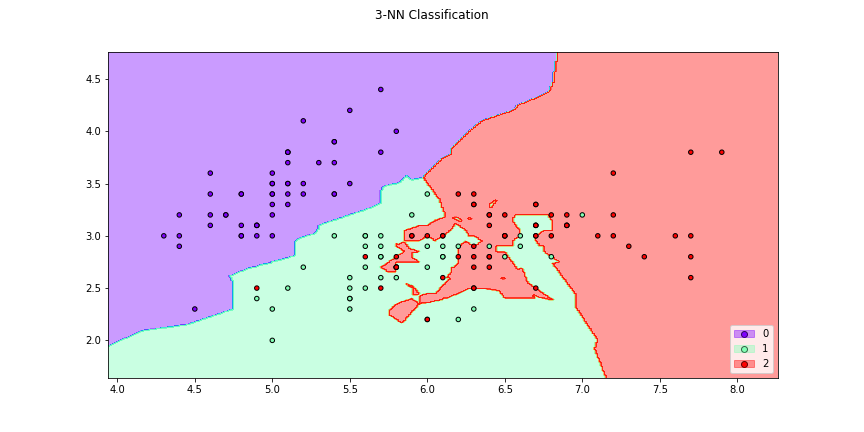
>>> # We can also make with the transformation determined by a metric,
>>> # a transformer or a DML Algorithm
>>> f3 = dml_plot(X[:,[0,1]],y,clf=ncmc,dml=nca,title = "NCMC Classification + NCA",
>>> cmap="rainbow",figsize=(12,6))
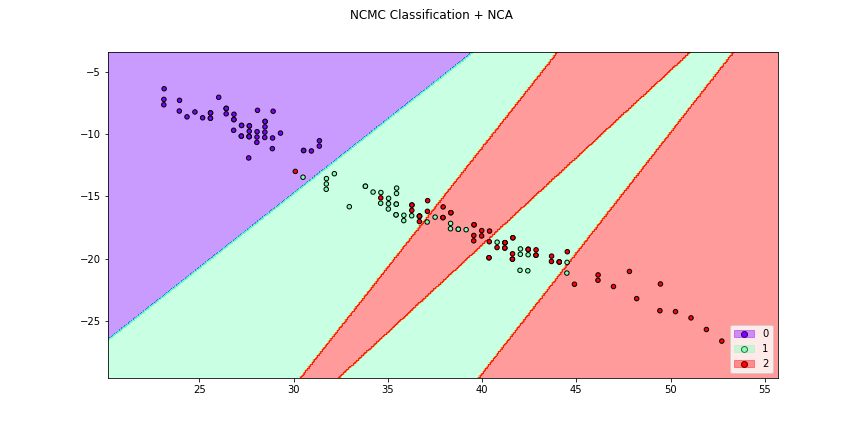
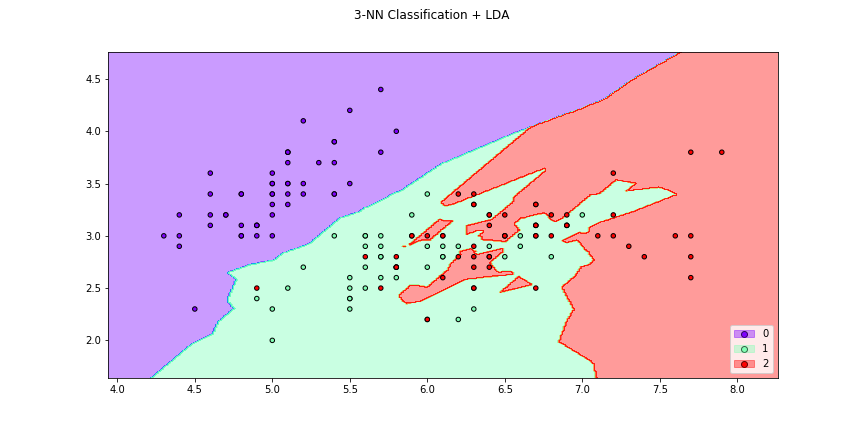
>>> f4 = knn_plot(X[:,[0,1]],y,k=2,dml=lda,title="3-NN Classification + LDA",
>>> cmap="rainbow",figsize=(12,6))
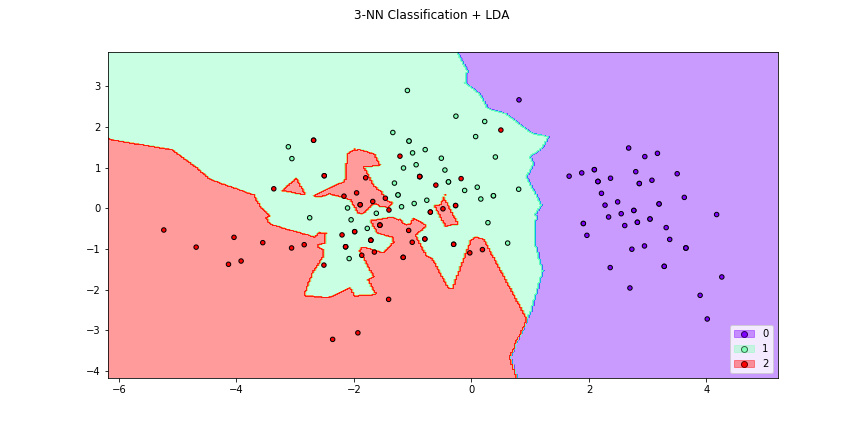
>>> # Or we can see how the distance changes the classifier region
>>> # using the option transform=False
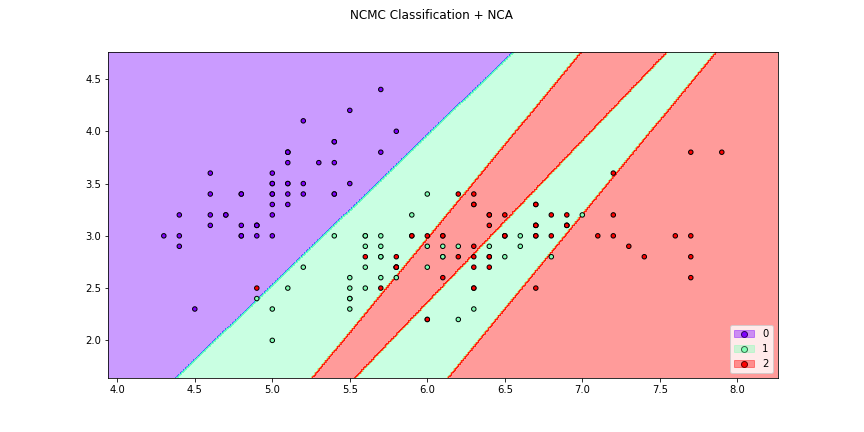
>>> f5 = dml_plot(X[:,[0,1]],y,clf=ncmc,dml=nca,title = "NCMC Classification + NCA",
>>> cmap="rainbow",transform=False,figsize=(12,6))
>>> f6 = knn_plot(X[:,[0,1]],y,k=2,dml=lda,title="3-NN Classification + LDA",
>>> cmap="rainbow",transform=False,figsize=(12,6))
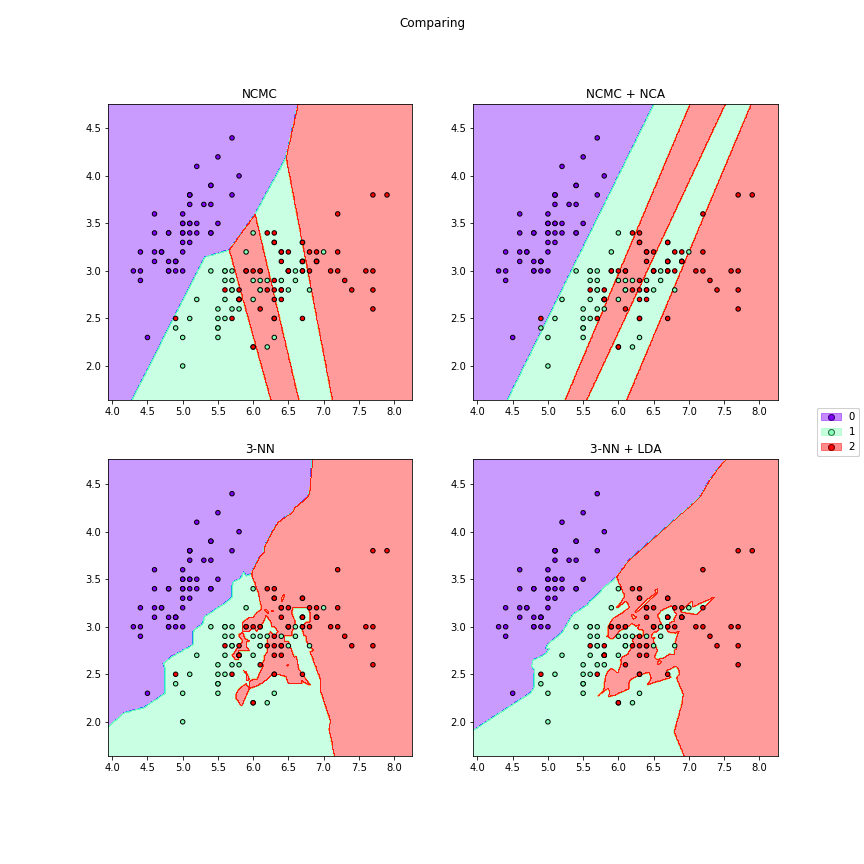
>>> # We can compare different algorithms or distances together in the same figure
>>> f7 = dml_multiplot(X[:,[0,1]],y,nrow=2,ncol=2,ks=[None,None,3,3],
>>> clfs=[ncmc,ncmc,None,None],dmls=[None,nca,None,lda],
>>> transforms=[False,False,False,False],title="Comparing",
>>> subtitles=["NCMC","NCMC + NCA","3-NN","3-NN + LDA"],
>>> cmap="rainbow",figsize=(12,12))
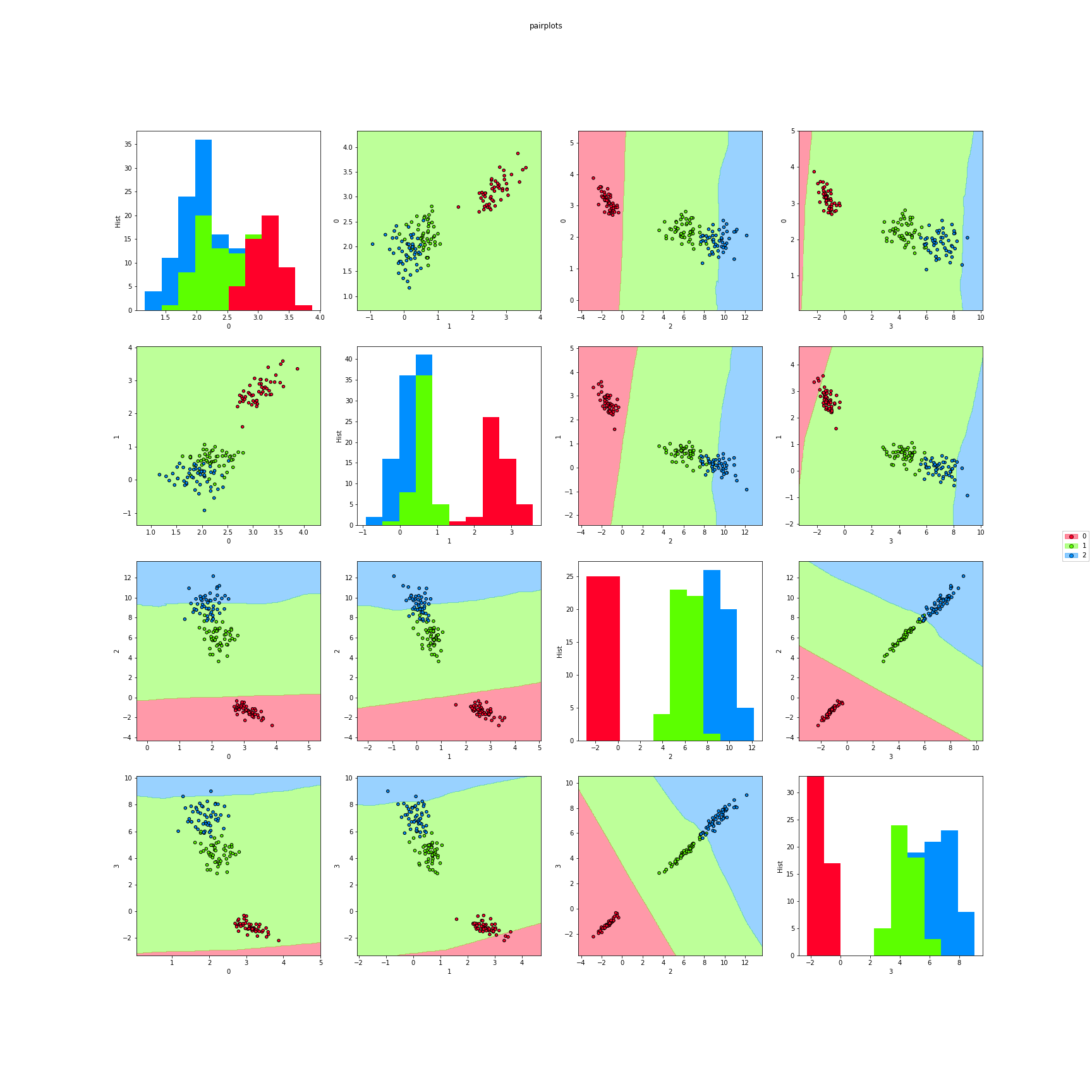
>>> # Finally, we can also plot each pair of attributes. Here the classifier region
>>> # is made taking a section in the features space.
>>> f8 = knn_pairplots(X,y,k=3,sections="mean",dml=nca,title="pairplots",
>>> cmap="gist_rainbow",figsize=(24,24))
Tuning parameters¶
>>> import numpy as np
>>> from sklearn.datasets import load_iris
>>> from dml import NCA, tune
>>> # Loading dataset
>>> iris = load_iris()
>>> X = iris['data']
>>> y = iris['target']
>>> # Using cross validation we can tune parameters for the DML algorithms.
>>> # Here, we tune the NCA algorithm, with a fixed parameter learning_rate='constant'.
>>> # The parameters we tune are num_dims and eta0.
>>> # The metrics we use are 3-NN and 5-NN scores, and the final expectance metadata of NCA.
>>> # A 5-fold cross validation is done twice, to obtain the results.
>>> results,best,nca_best,detailed = tune(NCA,X,y,dml_params={'learning_rate':'constant'},
>>> tune_args={'num_dims':[3,4],'eta0':[0.001,0.01,0.1]},
>>> metrics=[3,5,'final_expectance'],
>>> n_folds=5,n_reps=2,seed=28,verbose=True)
*** Tuning Case {'num_dims': 3, 'eta0': 0.001} ...
** FOLD 1
** FOLD 2
** FOLD 3
** FOLD 4
** FOLD 5
** FOLD 6
** FOLD 7
** FOLD 8
** FOLD 9
** FOLD 10
*** Tuning Case {'num_dims': 3, 'eta0': 0.01} ...
** FOLD 1
** FOLD 2
** FOLD 3
** FOLD 4
...
>>> # Now we can compare the results obtained for each case.
>>> results
3-NN 5-NN final_expectance
{'num_dims': 3, 'eta0': 0.001} 0.963333 0.970000 0.890105
{'num_dims': 3, 'eta0': 0.01} 0.966667 0.963333 0.916240
{'num_dims': 3, 'eta0': 0.1} 0.970000 0.963333 0.935243
{'num_dims': 4, 'eta0': 0.001} 0.956667 0.963333 0.897238
{'num_dims': 4, 'eta0': 0.01} 0.956667 0.963333 0.922415
{'num_dims': 4, 'eta0': 0.1} 0.960000 0.963333 0.947319
>>> # We can also take the best result (respect to the first metric).
>>> best
({'eta0': 0.1, 'num_dims': 3}, 0.97000000000000008)
>>> # We also obtain the best DML algorithm already constructed to be used.
>>> nca_best.fit(X,y)
>>> # If we want, we can look at the detailed results of cross validation for each case.
>>> detailed["{'num_dims': 3, 'eta0': 0.01}"]
3-NN 5-NN final_expectance
SPLIT 1 0.966667 0.966667 0.923293
SPLIT 2 0.966667 0.966667 0.922091
SPLIT 3 1.000000 0.966667 0.907416
SPLIT 4 0.966667 0.966667 0.903700
SPLIT 5 0.966667 0.966667 0.915030
SPLIT 6 0.966667 0.966667 0.905189
SPLIT 7 0.966667 0.966667 0.922051
SPLIT 8 0.933333 0.933333 0.933400
SPLIT 9 0.966667 1.000000 0.912236
SPLIT 10 0.966667 0.933333 0.917992
MEAN 0.966667 0.963333 0.916240
STD 0.014907 0.017951 0.008888